Research
In this lab, we study systems at the interface of biophysics and soft condensed matter. We are interested in exploring biologically-derived materials as tools to answer physics questions, and using physics-based inquiry to understand life and living systems. Many of the experiments in the lab explore the dynamics of large DNA molecules, which for many years have served as a model system to study the dynamics of polymers, using techniques such as optical fluorescence microscopy and microfluidics. Areas of physics relevant to our research include thermodynamics and statistical mechanics, as well as fluid dynamics.
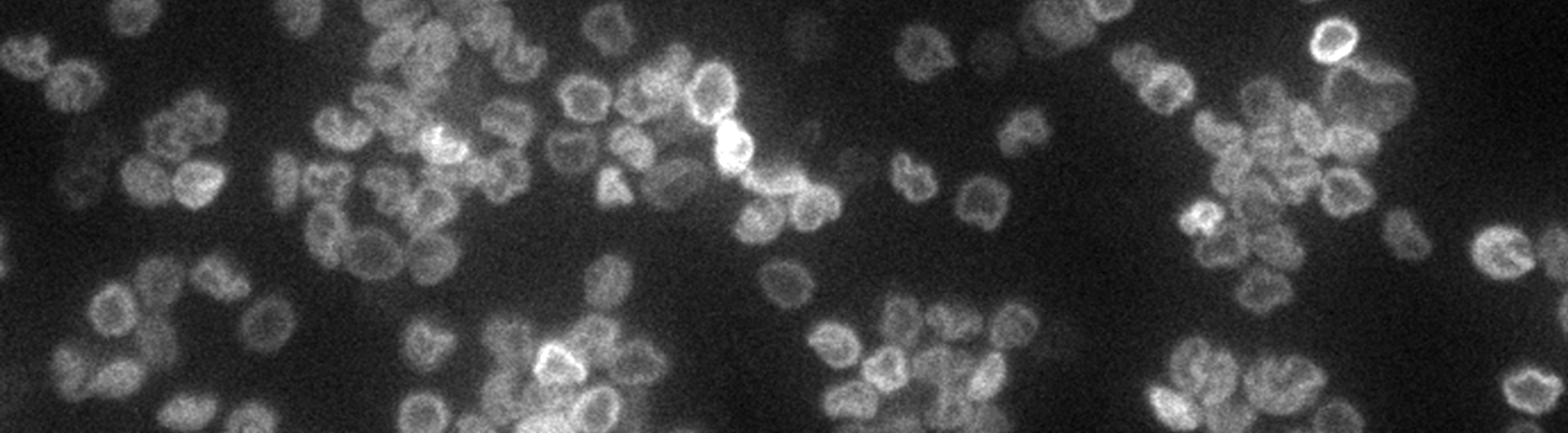
Molecular Chainmail: Physics of the Kinetoplast
Most of our DNA is in the form of long strands. In contrast, bacterial DNA is found in closed circular loops. There is a family of parasites called trypanosomes, which cause tropical diseases like Sleeping Sickness, whose mitochondrial DNA consists of thousands of loops linked together in a topological network like DNA chainmail. We are using kinetoplasts as model systems to investigate materials physics questions like "how do materials with non-covalent chemical bonds stretch?" and "how does a two dimensional polymer crumple?"
Complex Fluids and Non-Equilibrium Thermodynamics with Biopolymers
The interior of a living cell is a system far from equilibrium and consists of thousands of interacting components. When attempting to understand the physics of life, one must realize that living systems only reach thermal equilibrium upon death. However, even the simplest cell has hundreds of thousands of interacting components, quite complex by physics standards. We desire a system that embodies the thermodynamics and fluid mechanics of life without its complexity, and for that we will turn to active fluids: systems in which molecules are self-propelled by an internal supply of chemical energy, rather than diffusing around with Brownian motion. We are investigating the interaction of large DNA molecules with dense solutions of actin protein filaments, as a step towards a minimal system for understanding the physics of life.
Because we have just begun this project, please check out Stephen de Camp's website on active matter.
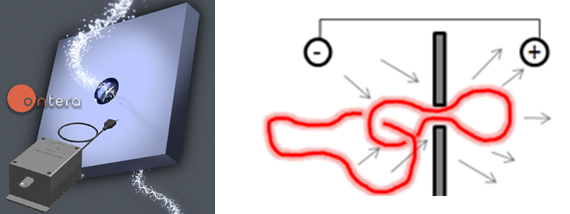
Nanopore Translocation
A nanopore is a new genetic sequencing technology that detects DNA by measuring an electrical current passing through a nanometer-scale hole in a membrane, so small that a DNA molecule is big enough to block the flow of ions. We are working with a startup company called Ontera to investigate the translocation of topology complex DNA molecules, such as linked ring systems that come from kinetoplasts.
Theory and Simulations
In addition to our experimental work, we develop theory and simulations and work with collaborators to better understand the systems we study in the lab and investigate other problems in statistical and soft matter physics. Our first computational study can be found on the Publications page.
Prior Work
Knots in DNA
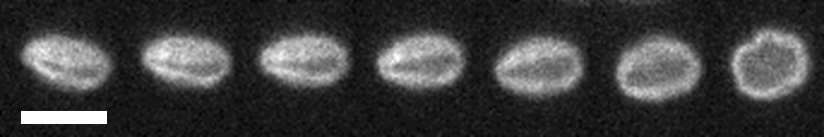
Many modern materials are made of polymers, long chain-like molecules that can become entangled with one-another. These entanglements affect the elastic and viscous properties of polymer materials, and in the Doyle Group at MIT I studied knots in DNA as a minimal version of polymer entanglement. Knots in DNA form naturally in cells, and can interfere with the function of genetic sequencing devices. My work focused on the effect of knots on the elasticity of individual molecules, and the processes by which knots can be controlled and removed with electric fields.
DNA Tetris
My Ph.D. work at McGill University used nanofluidic channels (very small tubes) to confine DNA molecules. I used devices that forced molecules to adopt configurations that looked like pieces from the video game Tetris, reducing the complexity of an individual molecule from thousands of degrees of freedom down to a number of discrete configurations that could be counted my hands. I used these devices to make measurements of how much energy is required to confine a polymer in a space much smaller than its natural size.